News
- August 4, 2020: Teaching about molecular orbitals for undergraduates: Developing a Virtual Reality Approach toward a Better Understanding of Coordination Chemistry and Molecular Orbitals by Ruxi Dai, et. al.
- February 4, 2020: Augmented reality video on coronavirus spikes seen in electron microscopy made with ChimeraX.
- November 20, 2019: Guide to recording mixed reality videos with ChimeraX and a depth sensing camera. Example video about opioids. November 4, 2019: Visualizing biomolecular electrostatics in virtual reality with UnityMol-APBS. Example acetylcholinesterase video and engineered enzyme video.
- October 16, 2019: UCSF story and video on Matt Jacobson and Beth Winger's use of VR to look leukemia drug resistance.
- October 10, 2019: Molecular virtual reality hands-on booth at UCSF Sharecase using ChimeraX.
- September 9-13, 2019: Virtual reality training for molecular structures and cryoEM at the Bioinformatics and Computational Biosciences Branch (BCBB) Training Seminars at Rocky Mountain Laboratories organized by Meghan McCarthy, Phil Cruz and Victor Kramer of Medical Science and Computing, Inc and Darrell Hurt of NIAID.
- September 4, 2019: Lenny Lupin-Jimenez at UCSF testing FCS-VR flow cytometery application developed by Tom Skillman of Immersive Science.
- August 26, 2019: UCSF professor Tom Ferrin discussed VR in drug discovery at the ACS annual meeting.
- August 8, 2019: Peppy: A Virtual Reality Environment for Exploring the Principles of Polypeptide Structure is an educational VR application for understanding protein secondary structure available at GitHub.
- July 24, 2019: VR walk-in workshop for light microscopy hosted by Jordan Briscoe at the Biological Imaging Development Colab at UCSF.
- July 3, 2019: Adam Frost lab at UCSF setup using Oculus Rift S headset to analyze high resolution cryoEM maps.
- June 26, 2019: Cancer drug resistance anaylzed by UCSF researchers assisted by ChimeraX virtual reality, described in ATP-competitive inhibitors midostaurin and avapritinib have distinct resistance profiles in exon 17-mutant KIT.
- June 20, 2019: Influenza vaccine research discussion on the PBS news hour with NIAID director Anthony Fauci including use of ChimeraX virtual reality to look at antibody binding sites.
- June 5, 2019: Max Krummel lab at UCSF setup using Vive Pro headset to analyze 3d lightsheet microscopy time series. Replaced Vive Pro with Oculus Rift S in July for better tracking in space with many glass windows.
-
May 2, 2019:
Three new PC VR headsets coming to market Spring or Summer 2019.
- Valve Index with Knuckles hand controllers using 87 sensors, and wider 130 degree field of view (review).
- Oculus Rift S with inside-out tracking, low cost ($400) with many small improvements (review).
- HP Reverb with higher resolution 2160 x 2160 pixels per eye (review)
- April 22, 2019: Dyche Mullins' lab at UCSF setup Vive Pro VR system, Windows 10, RTX 2080 graphics for 3d optical microscopy time series to study cell motility and actin dynamics.
- March 27, 2019: VRmol web VR app for molecular docking (using Vina) and examining genome mutations on PDB atomic models described at bioRxiv.
- February 26, 2019: ChimeraX VR video tutorials for medical imaging (details), molecular models, cryoEM maps, and a look at opiod drug binding.
- February 20, 2019: David Drubin and Georjana Barnes lab at UC Berkeley trying VR for lightsheet microscopy.
- February 19, 2019: Cell Toss VR app by Blair Lyon at the Allen Institute for Cell Science shows 3d optical microscopy of cells in different stages of mitosis.
- January 10, 2019: Yifan Cheng lab at UCSF setup MacBook Pro / eGPU / Vive Pro system for high resolution cryoEM maps.
- January 9, 2019: Dr Kurtis Auguste at UCSF Benioff Children's hospital shows patients VR tour of their brain before surgery to reduce anxieties.
- December 17, 2018: Matt Jacobson lab at UCSF setup iMac / Vive Pro system for studying drug binding.
- November 29, 2018: Loic Royer trying Vive Pro on microscopy data at CZ BioHub.
- November 20, 2018: Tests of VR on Windows laptops use with ChimeraX.
- November 15, 2018: How to setup VR on Mac laptop and desktop computers for use with ChimeraX.
- November 7-9, 2018: VR and AR science visualization hackathon in San Francisco hosted by Springer Nature. Sign up.
- October 26, 2018: How to setup VR on Linux for use with ChimeraX.
- October 23, 2018: Making Space: Molecules & Biomedical Imaging in Virtual Reality Open Swim a workshop to explore medical imaging data in VR at NC State University given by Meghan McCarthy and Phil Cruz.
- October 10, 2018: Opioids bound to mu opioid receptor ChimeraX VR demo.
- August 29, 2018: Nanome molecular VR for proteins and ligands. Extensive VR user interfaces. Free and commercial versions. Start-up company spun off from UC San Diego.
-
June 29, 2018:
Nano Simbox iMD
virtual reality interactive molecular dynamics developed by Interactive Scientific.
Sampling molecular conformations and dynamics in a multiuser virtual reality framework.
O'Connor M, Deeks HM, Dawn E, Metatla O, Roudaut A, Sutton M, Thomas LM, Glowacki BR, Sage R, Tew P, Wonnacott M, Bates P, Mulholland AJ, Glowacki DR.
Sci Adv. 2018 Jun 29;4(6) -
June 28, 2018:
ChimeraX VR article published describing drug binding site and light microscopy capabilities.
Molecular Visualization on the Holodeck.
Goddard TD, Brilliant AA, Skillman TL, Vergenz S, Tyrwhitt-Drake J, Meng EC, Ferrin TE.
J Mol Biol. 2018 Jun 28. pii: S0022-2836(18)30696-X -
June 24, 2018:
ConfocalVR article published describing 3D light microscopy visualization.
ConfocalVR: Immersive Visualization Applied to Confocal Microscopy.
Stefani C, Lacy-Hulbert A, Skillman T.
J Mol Biol. 2018 Jun 24. pii: S0022-2836(18)30664-8. - September 18, 2017: UCSF professors Derek Harmon and Kimberly Topp using VR to teach anatomy.
Research Applications
| 
| 
|
|
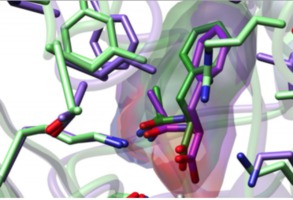
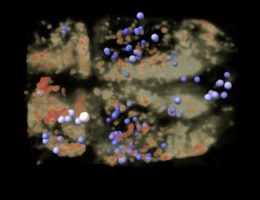
Drug binding sites Matt Jacobson lab Weekly group meetings using VR to analyze ligand binding using ChimeraX. Contact Wilian Cortopassi Coelho. |
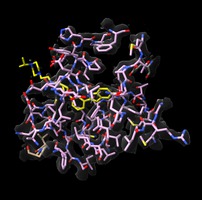
p97 inhibitors Michelle Arkin lab Inhibitors of p97 involved in protein recycling and possibly neurodegnerative disease. Virtual reality demonstration for UCSF Campaign. |
Antibiotic resistance Danica Fujimori lab Mechanism of post-translational methylation of a ribosome nucleotide causing antibiotic resistance. Shown in VR for Byer's award reception. |
|
|
|
|
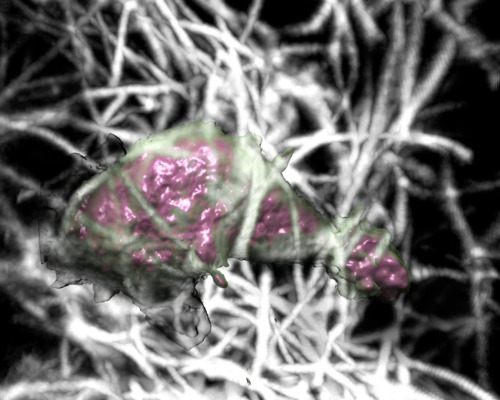
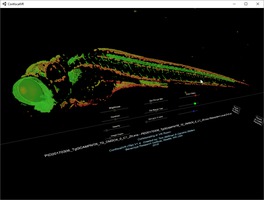
Cell motility 3D light microscopy Dyche Mullins lab ChimeraX VR displays 3D microscopy time series. Neutrophils crawling through collagen filaments. |
Calcium signaling in neurons Bo Huang lab Dan Xie observes calcium signaling in 8000 zebrafish neurons using 3d light-sheet microscopy time series. Demonstrated in VR for Byer's award lecture using ChimeraX VR. |
Neurodegeneration in zebrafish Steve Finkbeiner lab Neurodegeneration in zebrafish in 3d light microscopy time series from Jeremy Linsley at Glastone Institute shown with ConfocalVR. |

| 
|
|
DNA origami virtual lab
Shawn Douglas lab Interview with Shawn on the Foo Show about his VR nano-machine lab simulation. |
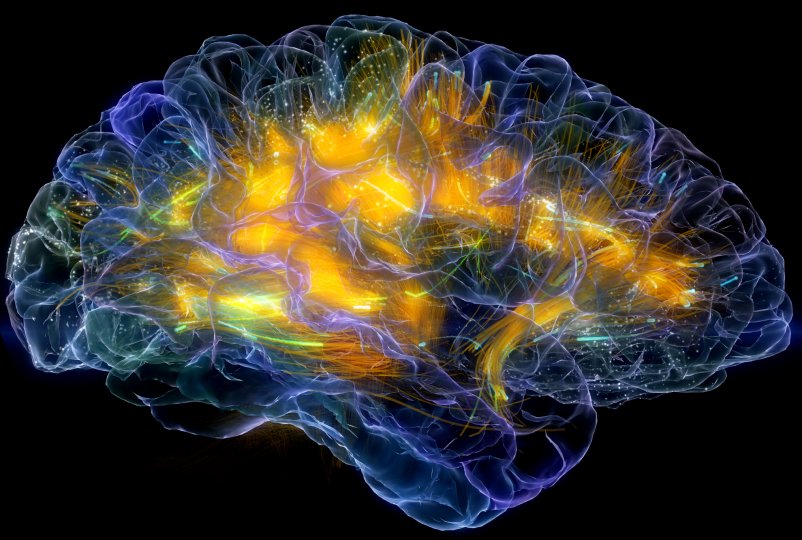
Brain disorder treatment Adam Gazzaley lab Neuroscape. |
VR Software for Research
|
|
|
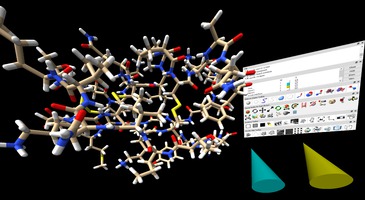
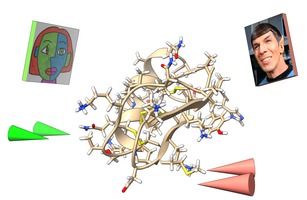
ChimeraX VR Tom Ferrin lab The ChimeraX visualization package can display atomic structures and microscopy data with the vr command. A tutorial explores mutations in a scorpion toxin. |
Collaborative structure viewing Tom Ferrin lab ChimeraX VR meeting command allows multiperson viewing of structures and microscopy data. |
|

|
|
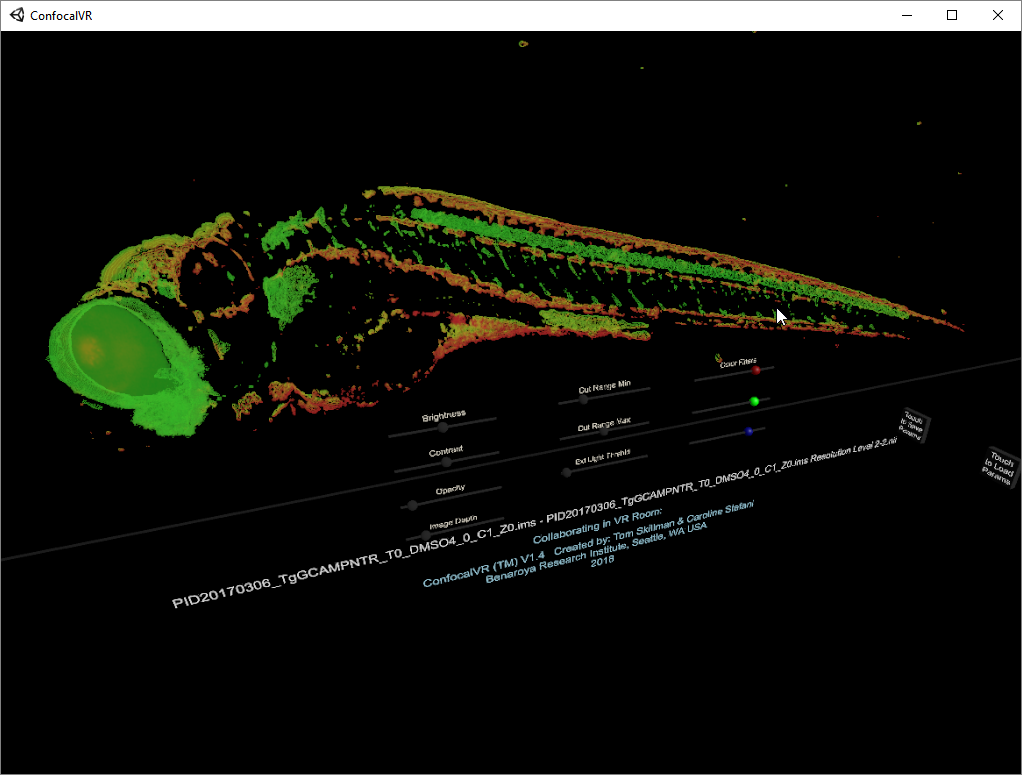
ConfocalVR Tom Skillman and Caroline Stefani at Benaroya Research Insitute ConfocalVR displays multichannel 3D light microscopy data. |
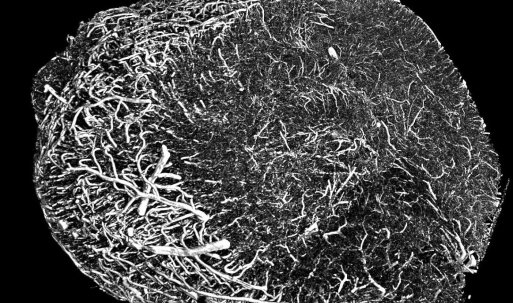
Protein Databank VR viewer Tom Skillman at Benaroya Research Institute AltPDB collaborative viewing of Protein Databank molecular structures. |

|
|
|
Nanome from Nanome, Inc. Nanome allows display and analysis of molecular structures of proteins including multiperson collaborative sessions. |
syGlass from IstoVizio, Inc. syGlass displays volumetric and microscopy data, designed for VR. |
Software for Education
|
|
|
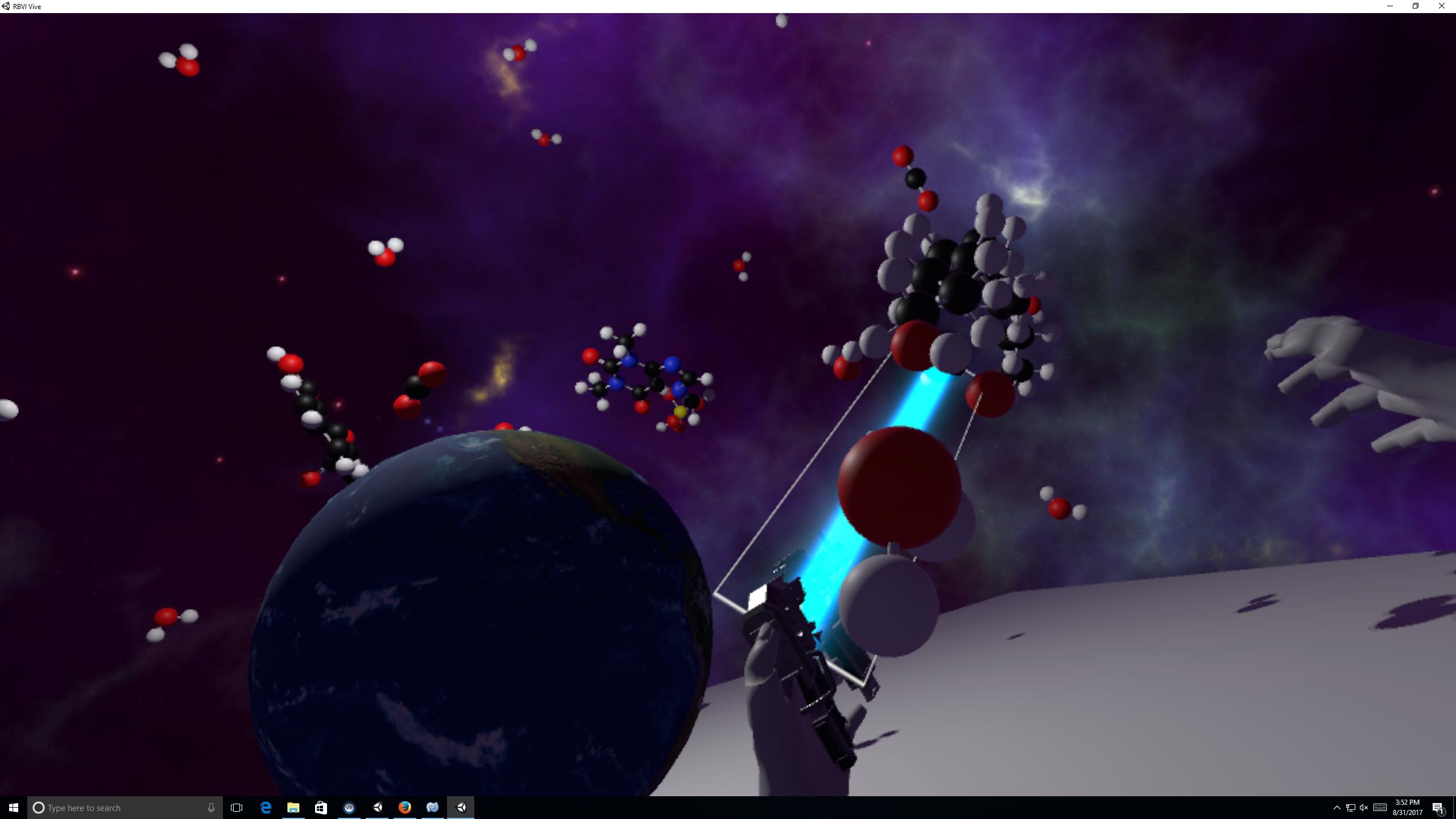
Molecular Zoo Alan Brilliant in Tom Ferrin lab Molecular Zoo is a biomolecule education VR program that lets children handle fully flexible molecules of life: water, aspirin, saturated fat, carbon dioxide, caffeine, ATP. |
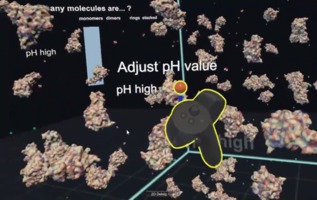
Peroxiredoxin Fish Tank David Doak at Norwich University of the Arts Nick Young and Bianca Haux at the Centre for eResearch at the University of Auckland Juliet Gerrard and Michael Barnett from the University of Auckland School of Biological Sciences Fishtank is a STEM education application showing the pH dependent formation of protein dimers, rings and tubes of an antioxidant enzyme. Currently (Sept 2018) only Unity source code releases are available at Github, but a binary release for Windows is coming. |

|
|
|
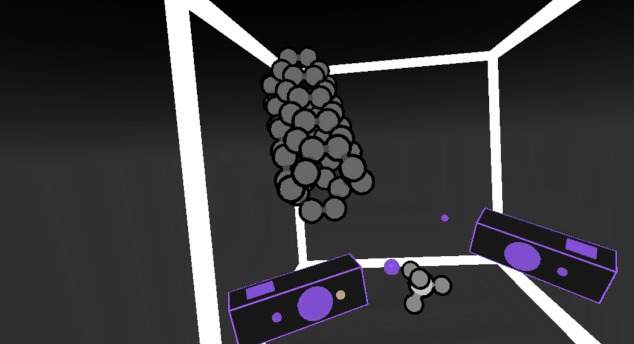
Protein Backbone Explorer David Doak at Norwich University of the Arts Work in progress: interactive polypeptide backbone viewer for teaching using Unity and Oculus SDK. Currently (Sept 2018) only Unity source code releases are available at Github, but a binary release for Windows is coming. |
Nano Simbox iMD Interactive Scientific Nano Simbox iMD allows exploring flexible molecules with molecular dynamics computed in real-time on a server. It has four scenarios, handling Buckminster fullerene, threading methane through a carbon nanotube, changing the screw-sense of an organic helicene molecule, and tying a knot in a 17 amino acid peptide. Developed by Interactive Scientific in collaboration with the Glowacki lab at University of Bristol, and described in this article. |

|
Water VR Jonas Boström and Magnus Norrby Water VR shows water in gas, liquid and solid phases for teaching chemisty, specifically aimed at 7th grade students. Developed by EduChemVR. |
VR Equipment

|
Shared VR equipment Tom Ferrin lab An HTC Vive virtual reality headset for viewing your molecular structures or microscopy data is available at the Visualization Vault in Genentech Hall at UCSF Mission Bay. Contact Tom Goddard. |
Headset Notes
We use Windows 10 Home on our VR computers. We use primarily SteamVR to interface to all headsets. Wikipedia headset comparison. VRcompare headset comparison.
- Vive Pro ($1400, released April 2018) - Higher resolution (1440x1600 per eye) makes reading text easier. More expensive ($1400 with hand-controllers and version 2 base-stations).
- Samsung Odyssey ($400, released Nov 2017) - Most portable, using inside-out tracking instead of base stations. Higher resolution (1440x1600 per eye). Shorter cord to computer limits range of motion. Hand controllers have no recharge port. Needs bluetooth used by controllers. Works with Windows Mixed Reality runtime, or SteamVR runtime.
- HTC Vive ($500 + $100 for headstrap, released April 2016) - Best tracking system. Deluxe headstrap is easier to adjust, more comfortable, and provides audio. Resolution (1080 x 1200 per eye).
- Oculus Rift ($400, released March 2016) - Best hand controllers, not as bulky, extra button, better button placement. Tracking is a pain, best to have a 3rd camera sensor, uses lots of USB ports. Resolution (1080 x 1200 per eye).
- Oculus DK2 - Historical developer kit 2 headset, released July 2014. Positional tracking using a single camera, loses tracking when facing away from camera. Resolution 960 x 1080 per eye. Frame rate 60-75 Hz.
- Oculus DK1 - Historical developer kit 1 headset, released March 2013. Only rotational tracking. No external base stations. Oculus SDK required app GPU code to correct lens distortion and chromatic aberration. Resolution 640 x 800 per eye. Frame rate 60 Hz.
Graphics Card Notes
We have extensive experience with the GPUs noted below and have also been experimenting with others, so this list may be a bit out of date:
- Nvidia GTX 1080 - Main card we have used at UCSF. No problems observed. 8 Gbytes of memory.
- Nvidia Quadro P6000 - Primarily because of its high price, we recommend this card only for special applications. We use this for both LCD shutter glasses and VR. The Quadro cards provide a 3-pin DIN connector for the left/right eye synchronization with shutter glasses. The IR emitter used with shutter glasses is overpowered by Vive (version 1) base stations so shutter glasses cannot be used with Vive base stations plugged in. This card has 4 display port outputs and no HDMI output. The Samsung Odyssey VR headset would not start using an HDMI to DisplayPort adapter with the Windows Mixed Reality setup program giving an error (Sep 2018). 24 Gbytes of memory. This card was generuously provided by NVIDIA Corporation's GPU grant program.
Projects
Virtual Reality in the San Francisco Public Libraries - pre-proposal to setup VR systems for informal STEM education in the 27 branches of the San Francisco public library. Full proposal due November 17, 2018.
Analysis of Molecules and Cells in Virtual Reality - Setup research VR systems in 5 UCSF labs for cryoEM structure refinement, 3D light microscopy, and multi-person VR collaborations. Funded Aug 1, 2018 for 1 year.
Exploring Drug Binding using Molecular Dynamics and Virtual Reality - proposal by Tom Goddard, Wilian Cortopassi Coelho, Matt Jacobson, Michael Grabe, Tristan Croll, and Tom Ferrin to study mutations effecting drug binding sites. Funded. January 2018.
Virtual Reality Molecular Playground - proposal for STEM education VR application to familiarize students with biomolecules. Not funded. January 2017.
People
Steven Abbott - Developer of many WebVR apps related to solvation.
Marc Baaden (Institut de Biologie Physico-Chimique) - Developer of UnityMol library for representing molecules in the Unity3D game engine.
Alan Brilliant (UCSC) - Developer of Molecular Zoo biomolecule education application.
Wilian Cortopassi Coelho (UCSF) - Studies drug-receptor interactions using VR in Matt Jacobson's lab. Publishing 3 drug studies in July 2018 using VR analysis.
Gareth Denyer (University of Sydney) - Developed Peppy protein structure teaching app with David Doak. Teaches structural biology at the Sydney Immersive learning laboratory.
David Doak (Norwich University of the Arts) - Developed peroxiredoxin fishtank STEM application and is working on a protein backbone explorer app.
Shawn Douglas (UCSF) - Developed VR lab simulation for DNA origami research.
Tom Ferrin (UCSF) - Provides shared HTC Vive VR equipment in the Visualization Vault.
Nicolas Floquet (U Montepellier) - Manages the MUSE-VR education facility in the Institut des Biomolécules Max Mousseron.
Christopher Fortney (NIH) - Manages the Virtual Reality Consulting Service at the NIH library.
Zack Gainsforth (Space Sciences Lab) - VR of electron microscopy of cometary dust.
Tom Goddard (UCSF) - ChimeraX VR developer.
Matt Jacobson (UCSF) - Analyzes drug binding, weekly use of VR in group meetings.
Meghan McCarthy (NIH) - Using ChimeraX VR with radiologists at NIH.
Dyche Mullins (UCSF) - VR visualization 3D light microscopy of crawling neutrophil cells.
Mike Schmid (Stanford-SLAC CryoEM Center) - VR visualization of EM tomography of bacteria, and atomic resolution virus capsids.
Tom Skillman (Immersive Science) - Founder of Immersive Science, developing VR science applications ConfocalVR for light microscopy, AltPDB for collaborative molecular structure viewing, and FCS-VR for flow cytometry analysis.
Caroline Stefani (Benaroya Research Institute) - Developer of ConfocalVR for light microscopy, working with Tom Skillman.
James Tyrwhitt-Drake - Developer of speech recognition command input from ChimeraX. Worked with Meghan McCarthy at NIH on virtual reality applications.
References
-
June 29, 2018:
Nano Simbox iMD
virtual reality interactive molecular dynamics developed by Interactive Scientific.
Sampling molecular conformations and dynamics in a multiuser virtual reality framework.
O'Connor M, Deeks HM, Dawn E, Metatla O, Roudaut A, Sutton M, Thomas LM, Glowacki BR, Sage R, Tew P, Wonnacott M, Bates P, Mulholland AJ, Glowacki DR.
Sci Adv. 2018 Jun 29;4(6) -
June 28, 2018:
ChimeraX VR article published describing drug binding site and light microscopy capabilities.
Molecular Visualization on the Holodeck.
Goddard TD, Brilliant AA, Skillman TL, Vergenz S, Tyrwhitt-Drake J, Meng EC, Ferrin TE.
J Mol Biol. 2018 Jun 28. pii: S0022-2836(18)30696-X -
June 24, 2018:
ConfocalVR article published describing 3D light microscopy visualization.
ConfocalVR: Immersive Visualization Applied to Confocal Microscopy.
Stefani C, Lacy-Hulbert A, Skillman T.
J Mol Biol. 2018 Jun 24. pii: S0022-2836(18)30664-8. -
June 23, 2018:
Review of web applications in chemistry and structural biology.
Towards Commodity, Web-Based Augmented Reality Applications for Research and Education in Chemistry and Structural Biology.
Luciano A. Abriata
arXiv:1806.08332 -
April 30, 2018: Nature Toolbox VR survey.
Virtual-reality applications give science a new dimension.
Matthews D.
Nature. 2018 May;557(7703):127-128. doi: 10.1038/d41586-018-04997-2. -
February 2018: Educational cell tour using EM segmentation.
Journey to the centre of the cell: Virtual reality immersion into scientific data.
Johnston APR, Rae J, Ariotti N, Bailey B, Lilja A, Webb R, Ferguson C, Maher S, Davis TP, Webb RI, McGhee J, Parton RG. Traffic. 2018 Feb;19(2):105-110. doi: 10.1111/tra.12538. Epub 2017 Nov 21.
Tom Goddard / UCSF / goddard@cgl.ucsf.edu